 When you think “genomics,” your mind probably pictures a human DNA strand. Well, my mind did, prior to meeting with Dr. Joseph Frassica and Dr. Felix Baader at HIMSS19 to discuss Philips’ approach to the tragic problem of healthcare-acquired infections that kill patients. Ever since that conversation, my mind’s eye is filled with images of MRSA cells like those shown here.
When you think “genomics,” your mind probably pictures a human DNA strand. Well, my mind did, prior to meeting with Dr. Joseph Frassica and Dr. Felix Baader at HIMSS19 to discuss Philips’ approach to the tragic problem of healthcare-acquired infections that kill patients. Ever since that conversation, my mind’s eye is filled with images of MRSA cells like those shown here.
At HIMSS19, Philips launched a solution that couples clinical informatics with genomic sequencing of bacteria to quickly identify and treat patients that are affected with tough-to-treat infections that, so often, result in death.
Healthcare-acquired infections (HAIs) are a worldwide challenge of epidemic, public health proportions. The World Health Organization recognizes that HAIs are a problem in both wealthy and developing countries. About 2.5 million Europeans die or are seriously debilitated each year from an HAI.
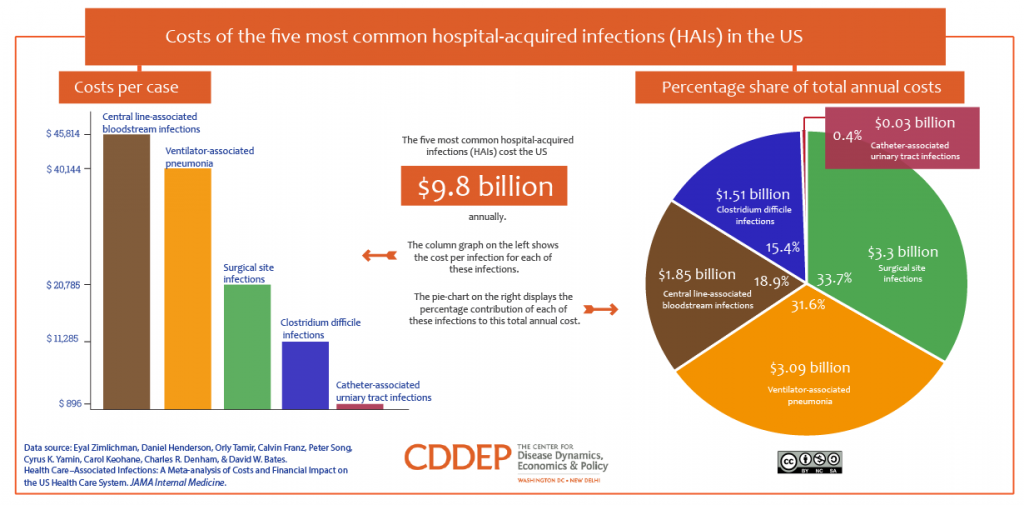
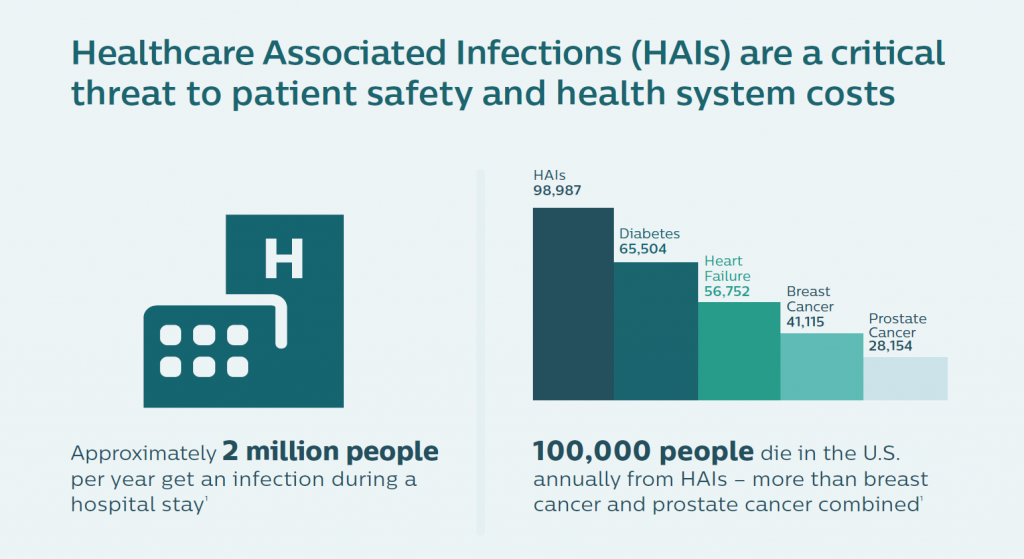
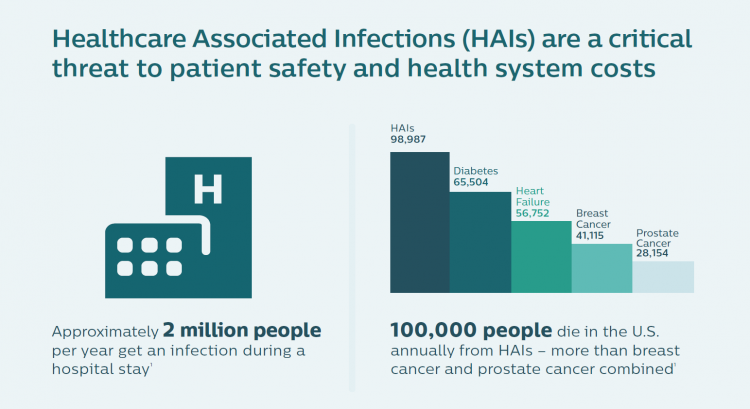
In the U.S., the five most common HAIs in the U.S. cost nearly $10 billion annually, according to an economic analysis published in JAMA Internal Medicine. More importantly, HAIs result in as many as 100,000 deaths a year in America. While the Leapfrog Group has recognized that U.S. hospitals have been working to improve the rate of nosocomial infections in America, there is a long way to go. One in 25 patients in U.S. hospitals still contracts an HAI, the Group’s recent report on HAIs attested.
Combating these deaths is a “winnable battle,” the U.S. Centers for Disease Control (CDC) believes. The lives that succumb to HAIs are cut short for reasons that are largely preventable. The researchers at Philips agreed with that sentiment, and worked to develop an approach to stem this crisis.
Philips’ solution marries informatics with genomics — leveraging two of the company’s key competencies. First, consider the informatics pillar, beginning with the many places and people where an infection can live: with a patient, a doctor, a nurse, a health aide; and, in an operating room, in an ICU, with a piece of surgical equipment, on an implement in the hospital cafeteria, for example. Now, digitally track that information through the patient’s journey during a hospital stay.
Second, deploy the skills to “genetically fingerprint pathogens,” the bacteria found in healthcare settings. It is now possible and increasingly cost-effective to do whole genome sequencing of pathogens in hospitals. The cost of doing so has fallen to the point where this approach to managing nosocomial outbreaks in large community hospitals is more feasible.
 For the research on which the eventual tool was based, the Philips team members worked collaboratively and globally, across time zones, with healthcare customers. I learned details about the research done in collaboration with the Westchester Medical Center in Valhalla, NY, an organization that has been firmly focused on reducing HAIs. Working iteratively on concepts over time, the project yielded solid proof points proving out the solution: the time to determine appropriate infection control action from an outbreak fell by 87%. Another clinical site involved with this research revealed many more clusters of infections detected with this approach than would otherwise not be identified. University of Massachusetts found 21 related clusters wiht the use of this technology, versus only one with current infection control practices.
For the research on which the eventual tool was based, the Philips team members worked collaboratively and globally, across time zones, with healthcare customers. I learned details about the research done in collaboration with the Westchester Medical Center in Valhalla, NY, an organization that has been firmly focused on reducing HAIs. Working iteratively on concepts over time, the project yielded solid proof points proving out the solution: the time to determine appropriate infection control action from an outbreak fell by 87%. Another clinical site involved with this research revealed many more clusters of infections detected with this approach than would otherwise not be identified. University of Massachusetts found 21 related clusters wiht the use of this technology, versus only one with current infection control practices.
“We build the family tree of the infections,” Dr. Baader explained. “If you have that, you know what the parent and siblings of each bacterium is. If there’s a parent and sibling, we can identify a transmission” of the bacteria to a patient or provider. Through machine learning and algorithms, using the data described in the informatics data flow, the clinical team can figure out whether the transmission was from a device, a patient in a shared room, or a caregiver.
The solution saves time and enhances workflow and productivity: typically for this process, it would take six practitioners to review the massive data set of 150,000 possible connections in the network between patients. The Philips IntelliSpace Epidemiology Solution narrowed this down to 40 transmission events by including pathogen genomics.
 “The interesting thing is that this sounds like rocket science to sequence a bacterium,” Dr. Frassica explained. “But that technology has progressed in the last few years. The sequencing step can be done for $80 today. Our pilot site at Westchester Valhalla sequenced each and every sample, demonstrating that the cost of sequencing for bacteria is less than the standard workflow cost of doing a bacterial culture.”
“The interesting thing is that this sounds like rocket science to sequence a bacterium,” Dr. Frassica explained. “But that technology has progressed in the last few years. The sequencing step can be done for $80 today. Our pilot site at Westchester Valhalla sequenced each and every sample, demonstrating that the cost of sequencing for bacteria is less than the standard workflow cost of doing a bacterial culture.”
Dr. Frassica, a pediatric intensivist by training, put this cost-effectiveness into a public health cost-benefit context. “There are more deaths from HAI infection than from breast and prostate cancer combined,” he noted, further saying that genome sequencing bacteria also allows the hospital to know when the situation is not an outbreak. Without the real-time information and insights, “You might be tempted to close the unit and move patients. In some cases, things that looked suspicious were not HAIs. They were genomically so far apart they couldn’t be acquired infections.” So the hospital management didn’t have to close beds, conserving that opportunity cost in those cases. Dr. Frassica shared a reference from a recent research article published in the New England Journal of Medicine to share more details about this phenomenon.
For more information, here’s Philips’ press release on the program.
Health Populi’s Hot Points: The public health and regional population health benefit to this informatics/genomics approach to managing HAIs is clear. Philips is taking a similar approach in oncology, announcing that development at HIMSS19 as well.
The public health implications are personal to me, having lost a family member to a runaway infection acquired in a hospital many years ago. I get the value of this tool from both the personal perspective and through my broader professional and public health economics lens.
There’s another aspect to learn from here, too, via the design approach taken by the Philips team. Dr. Frassica shared a “then” and “now” perspective: in the past decade, the team would, “ask a bunch of people what they needed,” translate that input into “what we thought they needed,” and then bring the client a solution.
“Now” for the HAI research, “we bring collaborators in for ideation and iteration throughout the process of creating the product,” Dr. Frassica described. “We iterated this product continuously with our partners” at Westchester and with other partner health care providers for feedback.
Why? “To make these products function in the real world,” Dr. Frassica asserted.
Such a co-creation strategy is the way forward to innovating technology that helps make health care better for both providers and patients — and save lives in the process.
Postscript — Today’s New York Times highlighted deaths from sepsis in U.S. hospitals, further highlighting the continued challenge to prevent avoidable deaths when possible. From the article: “My takeaway from this study is that we need to be even more vigilant to catch sepsis earlier in these vulnerable patients,” said Dr. Steven Simpson, medical director of the nonprofit Sepsis Alliance and a professor of pulmonary and critical care medicine at the University of Kansas. “People with cancer, heart failure, kidney disease and lung disease need to be informed: ‘Sepsis is your No. 1 enemy. These are the signs you need to be looking for.’”




 Thank you FeedSpot for
Thank you FeedSpot for